
Bonjour Hi! I am a first-year Computer Science Ph.D. student at McGill University and MILA - Québec AI Institute , advised by Prof. Mathieu Blanchette. Prior to this, I obtained my Master's degree from the same department in August 2024.
My research interests lie at the intersection of artificial intelligence and computational biology. Currently, I am focused on developing accurate and lightweight encoders to enhance the understanding of structure-function relationships in various biomolecules.
Aussi, je suis en train d'apprendre le français :)
Warning
Problem: The current name of your GitHub Pages repository ("Solution: Please consider renaming the repository to "
http://".
However, if the current repository name is intended, you can ignore this message by removing "{% include widgets/debug_repo_name.html %}" in index.html.
Action required
Problem: The current root path of this site is "baseurl ("_config.yml.
Solution: Please set the
baseurl in _config.yml to "Education
-

McGill University, MILA
School of Computer Science
Ph.D. Student Sep. 2024 - present -

McGill University
Master of Science in Computer Science Jan. 2023 - Aug. 2024
News
Selected Publications (view all )
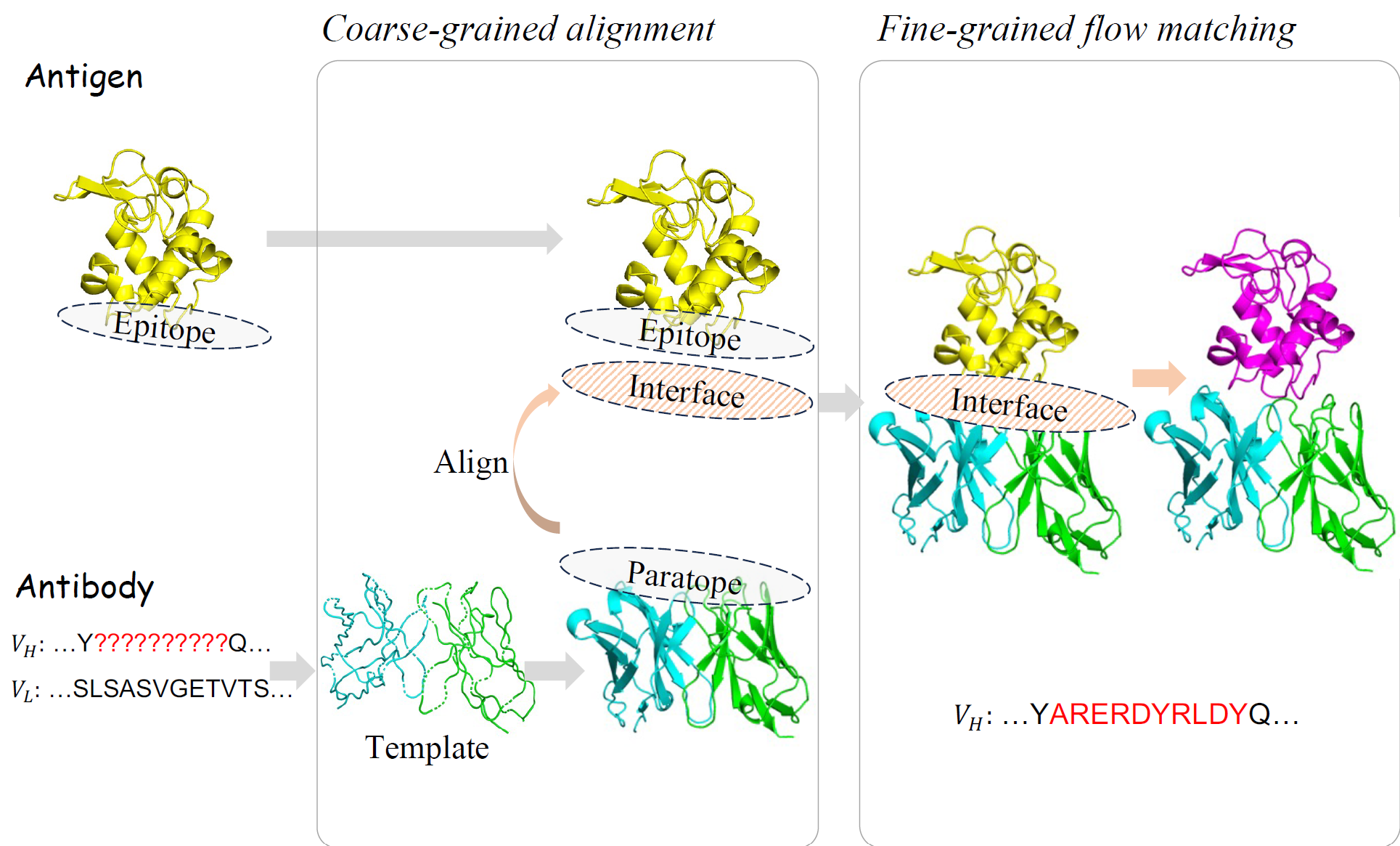
dyAb: Flow Matching for Flexible Antibody Design with AlphaFold-driven Pre-binding Antigen
Cheng Tan*, Yijie Zhang*, Zhangyang Gao*, Yufei Huang, Haitao Lin, Lirong Wu, Fandi Wu, Mathieu Blanchette#, Stan Z Li# (* equal contribution, # corresponding author)
The Thirty-Ninth AAAI Conference on Artificial Intelligence (AAAI) 2025 Oral Presentation
We introduce dyAb, a framework leveraging AlphaFold2-driven predictions and combining coarse-grained interface alignment with fine-grained flow matching to simulate dynamic interactions, significantly outperforming existing models and streamlining antibody design.
dyAb: Flow Matching for Flexible Antibody Design with AlphaFold-driven Pre-binding Antigen
Cheng Tan*, Yijie Zhang*, Zhangyang Gao*, Yufei Huang, Haitao Lin, Lirong Wu, Fandi Wu, Mathieu Blanchette#, Stan Z Li# (* equal contribution, # corresponding author)
The Thirty-Ninth AAAI Conference on Artificial Intelligence (AAAI) 2025 Oral Presentation
We introduce dyAb, a framework leveraging AlphaFold2-driven predictions and combining coarse-grained interface alignment with fine-grained flow matching to simulate dynamic interactions, significantly outperforming existing models and streamlining antibody design.
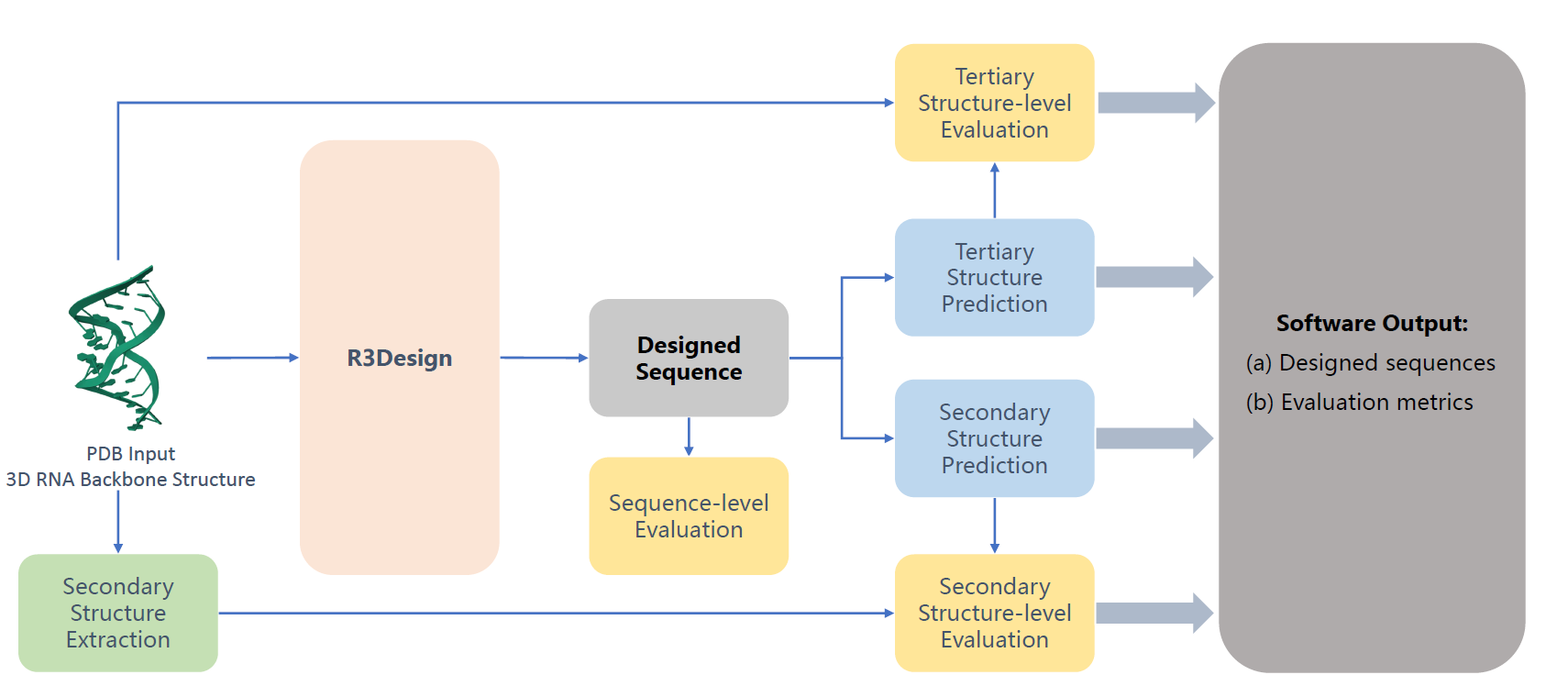
R3Design: Deep Tertiary Structure-based RNA Sequence Design and Beyond
Yijie Zhang*, Cheng Tan*, Zhangyang Gao*, Hanqun Cao, Siyuan Li, Siqi Ma, Mathieu Blanchette#, Stan Z Li# (* equal contribution, # corresponding author)
Briefings in Bioinformatics, 2025
R3Design is a tertiary structure-based RNA sequence design method that prioritizes tertiary interactions, significantly outperforming traditional secondary structure-based approaches. By enabling the design, folding, and evaluation of RNA sequences that fold into desired tertiary structures.
R3Design: Deep Tertiary Structure-based RNA Sequence Design and Beyond
Yijie Zhang*, Cheng Tan*, Zhangyang Gao*, Hanqun Cao, Siyuan Li, Siqi Ma, Mathieu Blanchette#, Stan Z Li# (* equal contribution, # corresponding author)
Briefings in Bioinformatics, 2025
R3Design is a tertiary structure-based RNA sequence design method that prioritizes tertiary interactions, significantly outperforming traditional secondary structure-based approaches. By enabling the design, folding, and evaluation of RNA sequences that fold into desired tertiary structures.
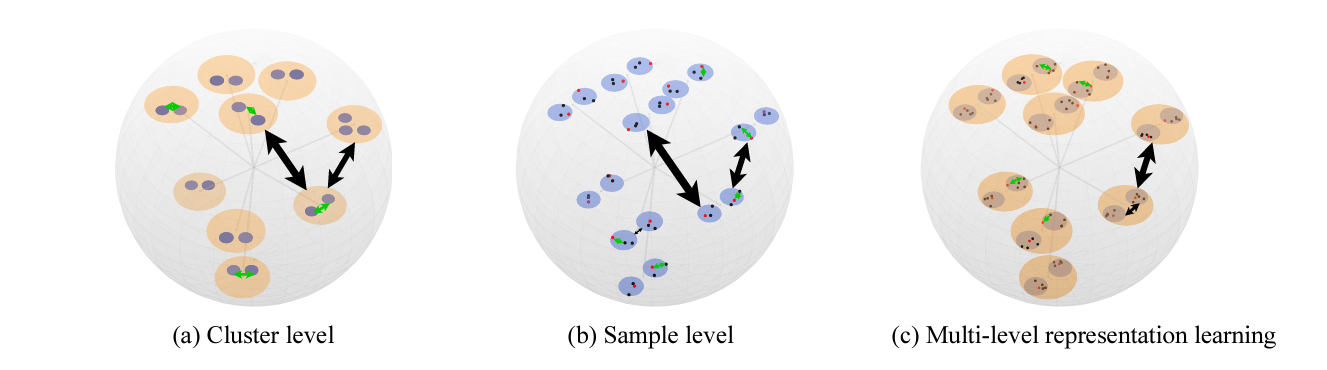
RDesign: Hierarchical Data-efficient Representation Learning for Tertiary Structure-based RNA Design
Cheng Tan*, Yijie Zhang*, Zhangyang Gao, Stan Z Li (* equal contribution)
The Twelfth International Conference on Learning Representations (ICLR) 2024
We proposed an RNA sequence design approach from the tertiary structure.
RDesign: Hierarchical Data-efficient Representation Learning for Tertiary Structure-based RNA Design
Cheng Tan*, Yijie Zhang*, Zhangyang Gao, Stan Z Li (* equal contribution)
The Twelfth International Conference on Learning Representations (ICLR) 2024
We proposed an RNA sequence design approach from the tertiary structure.
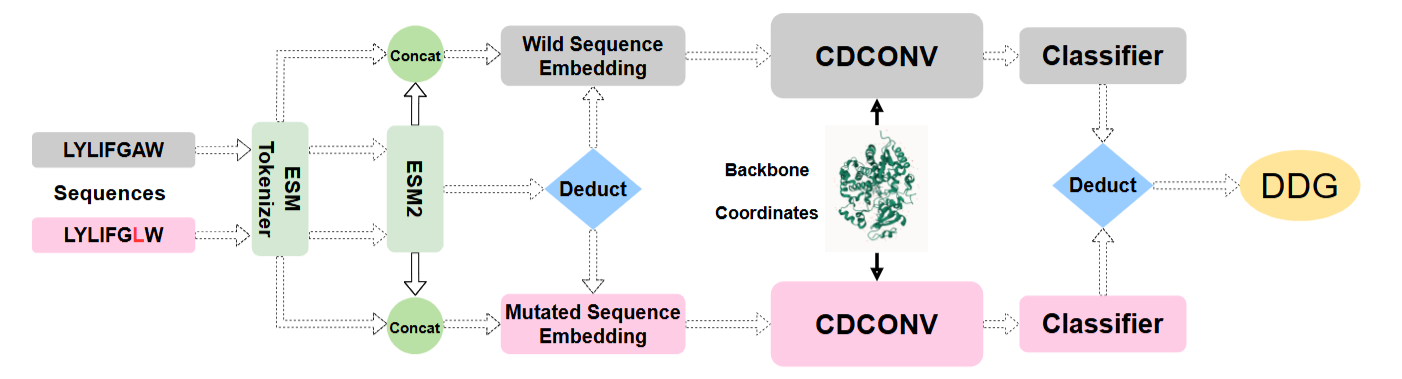
Efficiently Predicting Protein Stability Changes Upon Single-point Mutation with Large Language Models
Yijie Zhang, Zhangyang Gao, Cheng Tan, Stan Z. Li# (# corresponding author)
Arxiv 2023
Predicting protein stability changes from single-point mutations remains a significant challenge, hindered by feature extraction complexity and limited experimental data. Leveraging ESM-based large language models, we propose an efficient method integrating sequence and structural features to predict thermostability changes, supported by a carefully curated dataset to ensure fair model comparison.
Efficiently Predicting Protein Stability Changes Upon Single-point Mutation with Large Language Models
Yijie Zhang, Zhangyang Gao, Cheng Tan, Stan Z. Li# (# corresponding author)
Arxiv 2023
Predicting protein stability changes from single-point mutations remains a significant challenge, hindered by feature extraction complexity and limited experimental data. Leveraging ESM-based large language models, we propose an efficient method integrating sequence and structural features to predict thermostability changes, supported by a carefully curated dataset to ensure fair model comparison.